RNA thermometers
Detection of thermo-sensing RNA structures with Lead-Seq
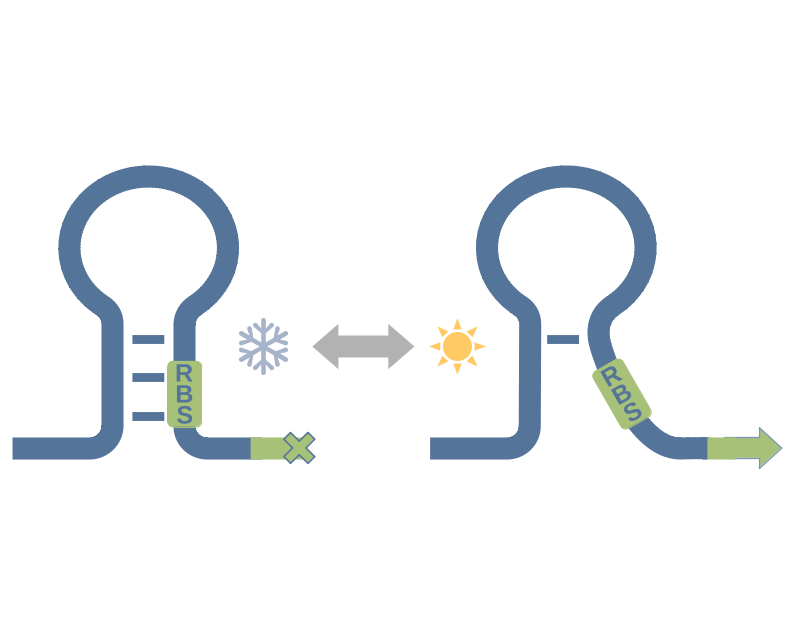
RNA thermometers are temperature sensing structures, typically located at the 5’-end of RNAs. They impede translation at low temperatures by forming a stable secondary structure and blocking the ribosome binding site and the start codon. At high temperatures, they melt open, reveal the ribosome binding site, and the translation can take place.
RNA thermometers can be identified with Lead-Seq, as shown in Twittenhoff, Brandenburg and Righetti et al., 2020. Code and Data for the RNA thermometer identification with Lead-Seq are available in GitHub.

To identify RNA thermometers from Lead-Seq data, Δscores are calculated from the distance between Lead-Scores at different temperatures. The Δscores around the ribosome binding site are compared to the Δscores of the whole transcript via Mann-Whitney-U test.


The Δscores and p-values from the Mann-Whitney-U tests around ribosome binding sites in Yersinia pseudotuberculosis are compared. About 80 transcripts have a negative Δscore and a significant p-value (green), indicating significant melting at higher temperatures. These 80 transcripts are good candidates for RNA thermometers. Of these, 3 are verified RNA thermometers from Righetti et al., 2016 (purple), and 2 are previously unknown RNA thermometers, verified in Twittenhoff, Brandenburg and Righetti et al., 2020 (pink).