Lead-Seq
Transcriptome-wide RNA structure probing with lead(II) ions.
Lead-seq is a structure probing method for bacteria, which combines chemical structure probing with high-throughput sequencing. It allows for the probing of RNA structures of entire transcriptomes at different conditions. The method was first published by Twittenhoff, Brandenburg and Righetti et al. in 2020, which was also reported in ScienceDaily. Code and Data for Lead-Seq are available in GitHub.

For Lead-Seq, a bacterial culture is treated with lead(II)-acetate. Lead(II) ions induce strand breaks in unpaired RNA regions. A second culture serves as negative control. Reverse Transcripase is added to both cultures, which creates a DNA copy of each RNA. In the lead-treated sample, the transcription stops when a lead-induced strand break is reached, whereas the transcription continues over this position in the control sample. Both samples are sequenced and mapped to the reference genome. The strand breaks are counted, and the normalized counts from both samples are used to calculate Lead-Scores. The Lead-Scores indicate single-stranded RNA regions.
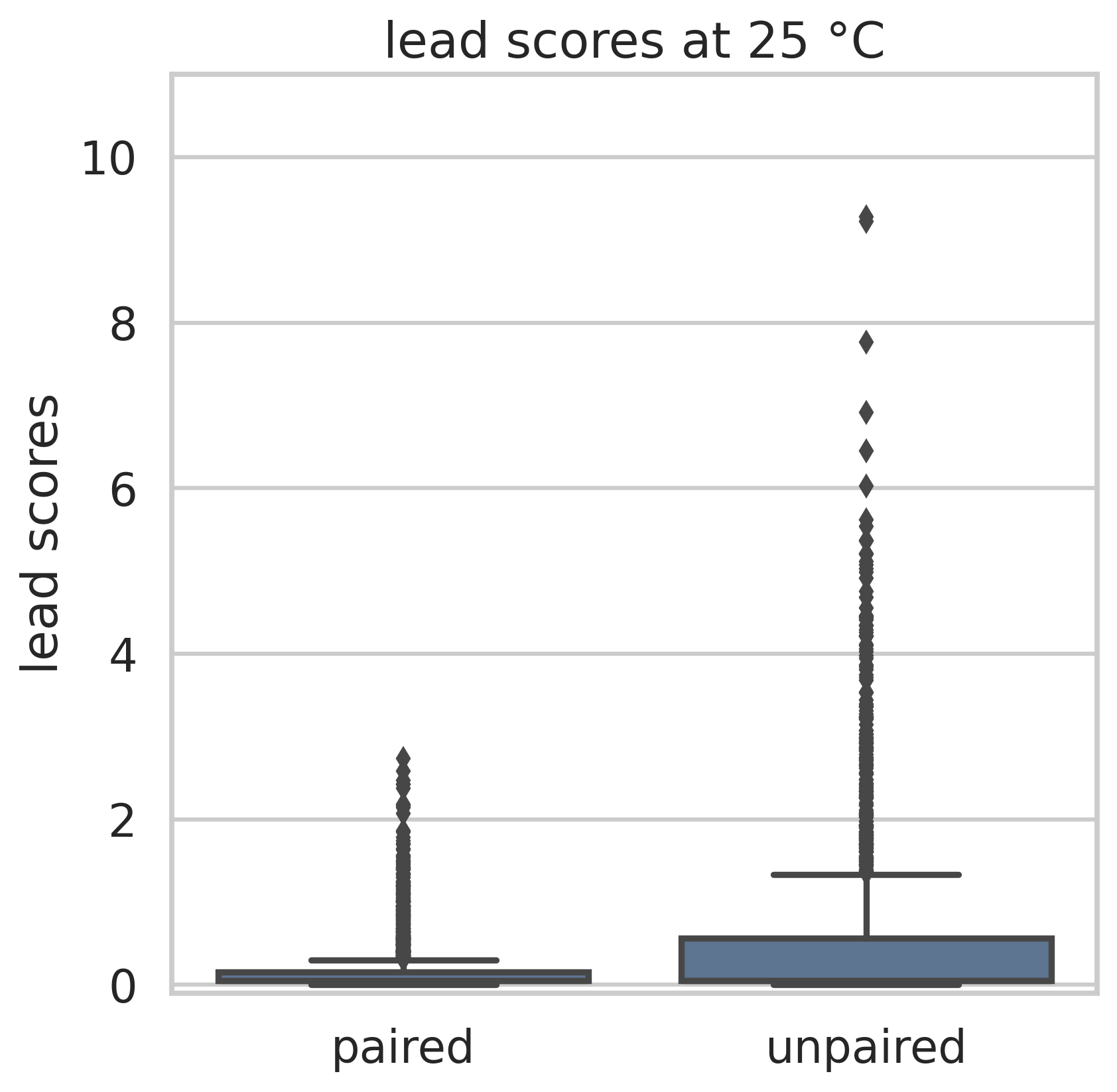
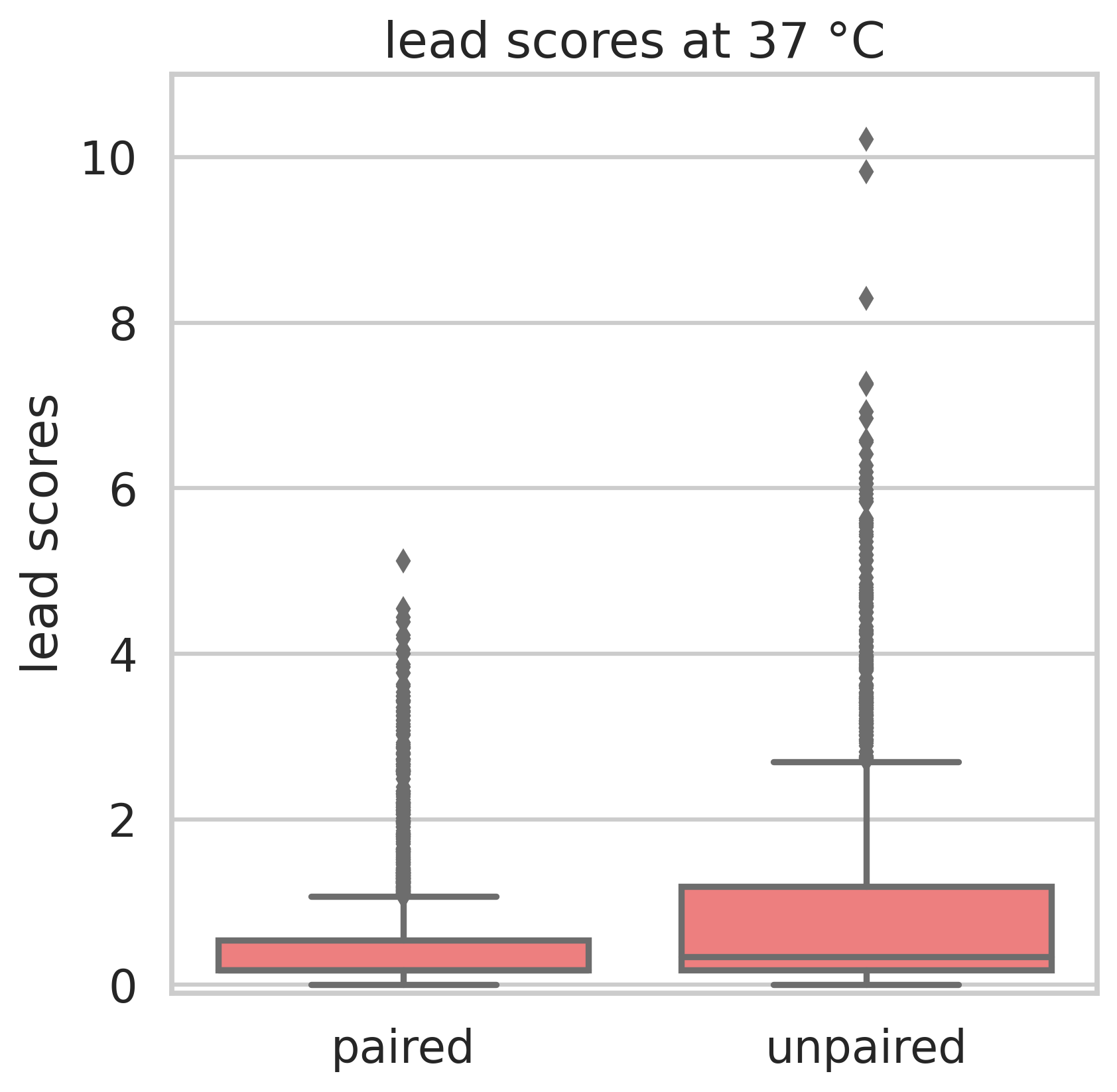

Lead-Seq was tested on the γ-proteobacterium Yersinia pseudotuberculosis at 25 °C and 37 °C and validated with tRNA structures. At both temperatures, the Lead-Scores are significantly higher at unpaired than at paired regions. The selectivity above 0.7 shows that Lead-Scores indicate unpaired nucleotides at different testing conditions.